Genetic diversity, population structure, and relationships among wild and domesticated almond (Prunus spp.) germplasms revealed by ISSR markers
Published 2020-06-25
Keywords
- cluster analysis,
- gene diversity,
- gene flow,
- population structure,
- wild almond
How to Cite
Abstract
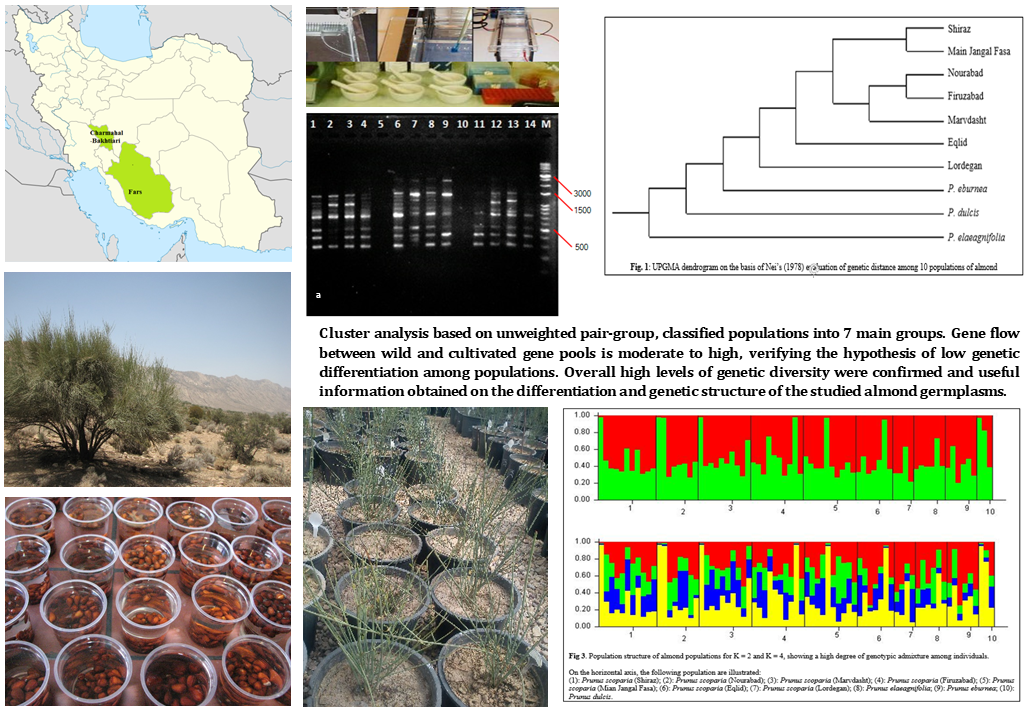
The use of diverse almond genetic resources to expand the genetic bases of commercial cultivars is important for almond breeders. Iran is within the center of origin for almond and enjoys a huge diversity of wild species and local cultivars of this important nut crop. Despite some reports, there is still a critical need to collect comprehensive information on the genetic diversity of almond germplasm in Iran. This study was conducted to evaluate the genetic diversity, structure, and relationships among a total of 75 individuals from 10 populations of 4 wild and cultivated almond species by using 12 inter-simple sequence repeat (ISSR) primer pairs. A total number of 353 DNA fragments were obtained of which 352 were polymorphic (99.69%). The average of polymorphism information content (PIC), marker index (MI), and resolving power (Rp) were 0.932, 27.211, and 7.882, respectively which indicated high discriminatory power of markers. Gene flow between wild and cultivated gene pools is shown to be moderate to high (Nm = 2.7607), which verifies the hypothesis of low genetic differentiation among populations. Cluster analysis based on unweighted pair-group, classified individuals into 7 major gene pools which showed the entire provenances were divided into 7 main groups. Overall high levels of genetic diversity were confirmed and useful information obtained on the differentiation and genetic structure of the studied almond germplasms. Future evaluation on morphological and physiological aspects, is necessary to identify the most promising individuals to be used directly in afforestation, landscape development as well as nut and oil production or indirectly in future almond and stone fruits breeding programs.





