Published 2020-10-26
Keywords
- Cucumis sativus,
- PCR,
- RFLP,
- 16SrVI-A,
- 16SrXII-A
How to Cite
Funding data
-
Ministry of Science Research and Technology
Grant numbers 2-64-16-94189
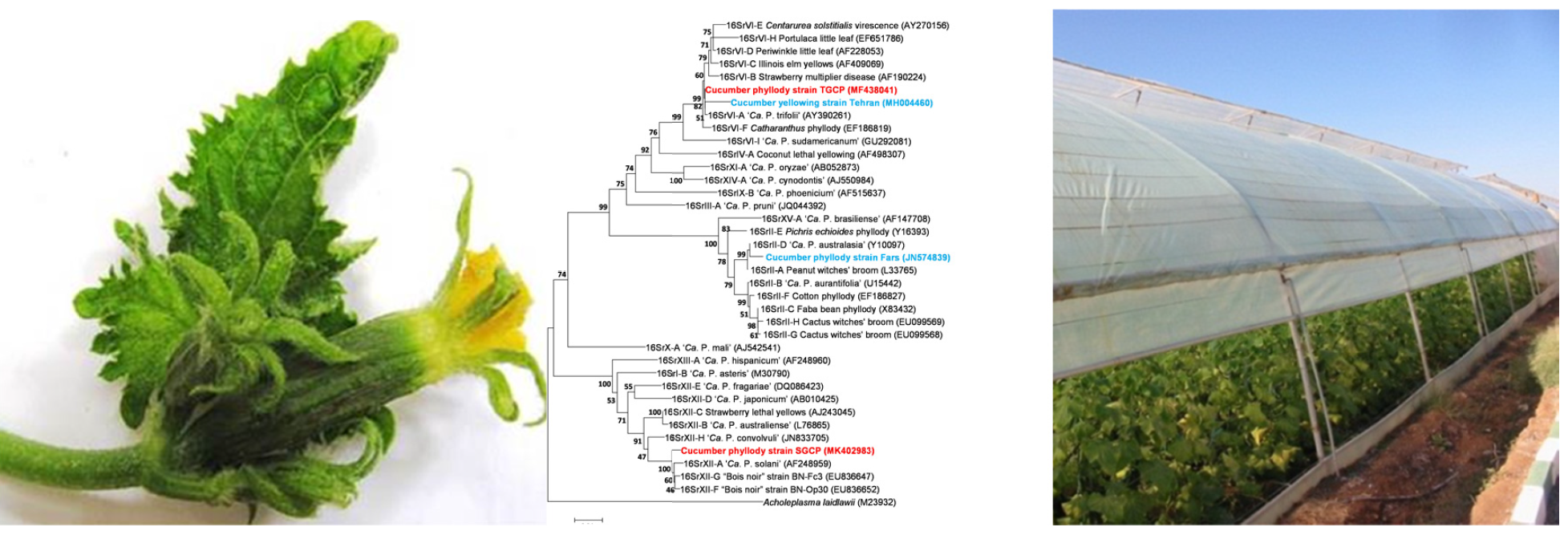
Abstract
Cucumber phyllody symptoms were observed in greenhouse cucumber plants during 20142018 in all surveyed areas of central and west of Iran where the highest disease incidence was up to 82% in Taft (Yazd province). Symptoms exhibited by diseased plants were virescence, phyllody and sterility of the flowers. For verification of phytoplasma presence and identity, total DNAs were extracted from 44 symptomatic and six asymptomatic plants that were subjected to PCR amplifying 16S rRNA genes of phytoplasmas. PCR amplicons of the expected size were obtained only from the symptomatic plants. RFLP analysis of R16F2n/R2 amplicons showed patterns identical to those of the clover proliferation (16SrVI) and “stolbur” (16SrXII) phytoplasma groups. Consensus sequences corresponding to phytoplasma strains from the two localities Taft and Shahrekord showed 99% identity with phytoplasmas enclosed in groups 16SrVI and 16SrXII, respectively. Phylogenetic analysis confirmed that these phytoplasmas cluster with ‘Candidatus Phytoplasma trifolii’ and ‘Ca. P. solani’, respectively. Virtual RFLP provided profiles identical to the patterns of 16SrXIIA and 16SrVIA phytoplasma subgroups. These phytoplasma subgroups were previously reported in different plant species growing near to the greenhouse cucumber areas in Iran, and play a possible role in the epidemiology of disease for its dissemination.





