Published 2025-05-28
Keywords
- Genome Association and Prediction Integrated Tool (GAPIT),
- intragenic molecular markers,
- Hierfstat,
- Musa textilis Nee,
- intragenic molecular markers
- population structure,
- Musa textilis Nee,
- population structure ...More
How to Cite
Copyright (c) 2025 Ms. Mariecris Rizalyn R. Mendoza, Dr. Antonio C. Laurena, Dr. Maria Genaleen Q. Diaz, Dr. Eureka Teresa Ocampo, Dr. Tonette P. Laude, Dr. Antonio G. Lalusin

This work is licensed under a Creative Commons Attribution 4.0 International License.
Abstract
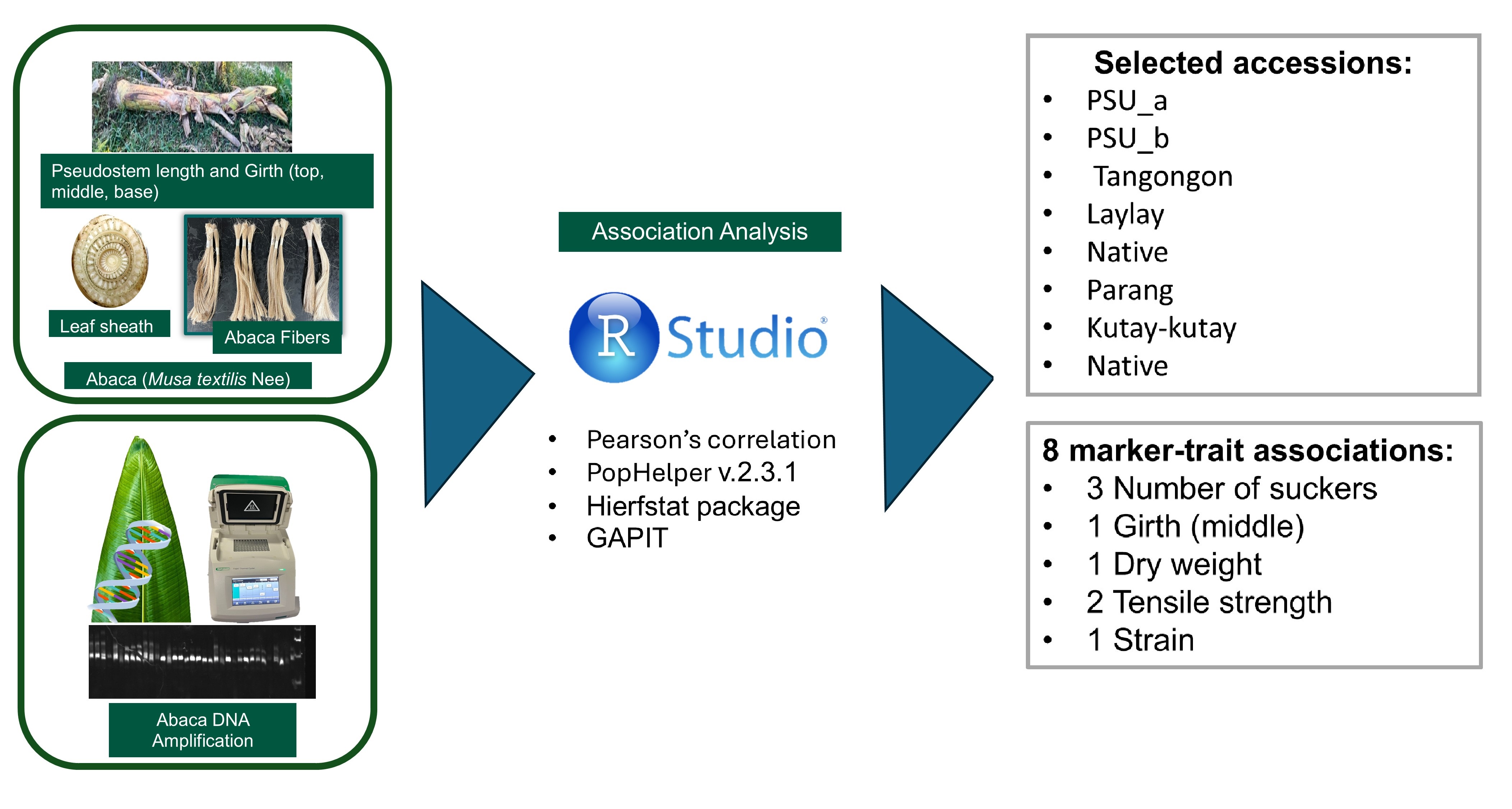
Abaca are leaf-fiber plants found predominantly in the Philippines. Our country holds most of the Manila hemp market, but the unknown genetic architecture of the fiber hinders the crop’s improvement. We developed intragenic molecular markers from genes related to fiber development and linked them to abaca fiber quality, with the goal of increasing precision of breeding. Pearson’s correlation package of the R programming software revealed a high positive relationship between the pseudostem’s top and middle girth (r=0.91), while a low negative correlation between the percent fiber percent fiber strain and the number of suckers (r= -0.42). The analysis also showed that the ultimate tensile strength was highly correlated with percent fiber percent fiber strain (r=0.33) and dry weight (r=0.34). Three subpopulations were determined using the STRUCTURE software, while Hierfstat computed an average 0.0648 Fst value, indicating moderate genetic diversity. Eight significant marker-trait associations (p-value <0.005) were identified with positive effects and >0.6% phenotypic variance explained (PVE). Eight markers from the COBRA-like protein, expansin, cellulose synthase, and auxin gene families were identified as linked to fiber quality and tensile strength. Our study identified nine abaca accessions with the trait of interest and the candidate genes. The significant molecular markers will be used to identify the hybrids with good fiber quality.
References
- ALSALEH A., 2022 - SSR-based genome-wide association study in Turkish durum wheat germplasms revealed novel QTL of accumulated platinum. - Mol. Biol. Rep., 49: 11289-11300. DOI: https://doi.org/10.1007/s11033-022-07720-7
- ALVES FIDELIS M.E., PEREIRA T.V.C., GOMES O.D.F.M., DE ANDRADE SILVA F., TOLEDO FILHO R.D., 2013 - The effect of fiber morphology on the tensile strength of natural fibers. - J. Mat. Res. Technol., 2(2): 149-157. DOI: https://doi.org/10.1016/j.jmrt.2013.02.003
- ASIS M., 2017 - GMO cotton could prompt renaissance of Philippine cotton industry. - https://allianceforscience.cornell.edu/blog/2017/11/gmo-cotton-could-prompt-renaissance-of-philippine-cotton-industry/.
- BAO Y., ZOU Y., HUANG X., REHMAN M., LIU C., SHI S., PENG D., FAHAD S., WANG B., 2024 - Genome-wide analysis and expression characteristics of small auxin-up RNAs genes in flax (Linum usitatissimum L.). - Industrial Crops Products, 217: 118874. DOI: https://doi.org/10.1016/j.indcrop.2024.118874
- BARRETO F.Z., ROSA J.R.B.F., BALSALOBRE T.W.A., PASTINA M.M., SILVA R.R., HOFFMANN H.P., DE SOUZA A.P., GARCIA A.A.F., CARNEIRO M.S., 2019 - A genome-wide association study identified loci for yield component traits in sugarcane (Saccharum spp.). - PLoS ONE, 14(7): 1-22. DOI: https://doi.org/10.1371/journal.pone.0219843
- BAYTAR A.A., PEYNIRCIOĞLU C., SEZENER V., FRARY A., DOĞANLAR S., 2022 - Association analysis of germination level cold stress tolerance and candidate gene identification in Upland cotton (Gossypium hirsutum L.). - Physiol. Mol. Biol. Plants, 28(5): 1049-1060. DOI: https://doi.org/10.1007/s12298-022-01184-6
- BEECKMAN T., PRZEMECK G.K., STAMATIOU G., LAU R., TERRYN N., DE RYCKEINZÉ RD, BERLETH T., 2002 - Genetic complexity of cellulose synthase a gene function in Arabidopsis embryogenesis. - Plant Physiol., 130(4): 1883-1893. DOI: https://doi.org/10.1104/pp.102.010603
- BEMILLER J., 2007 - Cell walls: economic significance, pp. 283-286. - In: ROBERTS K. (ed.) Handbook of plant science. John Wiley and Sons Ltd., New York, USA, pp. 1676.
- BETANCUR L., SINGH B., RAPP R.A., WENDEL J.F., MARKS M.D., ROBERTS A.W., HAIGLER C.H., 2010 - Phylogenetically distinct cellulose synthase genes support secondary wall thickening in Arabidopsis shoot trichomes and cotton fiber. - J. Integrative Plant Biol., 52(2): 205-220. DOI: https://doi.org/10.1111/j.1744-7909.2010.00934.x
- BOGUERO A.P., PARDUCHO M.A., MENDOZA M.R.DR, ABUSTAN MAM, LALUSIN AG., 2016 - Molecular screening of abaca (M. textilis Nee) accessions using microsatellite markers associated with resistance to bunchy top disease. - Philippine J. Crop Sci., 41(2): 13-19.
- BONDAD A.A., BADER R.F., TABORA P.C., 1979 - N, P, and K fertilizer on abaca. - National Science Development Board, NSDB Technology J., (Philippines), 4(3): 47-52.
- BUYYARAPU R., KANTETY R.V., YU J.Z., SAHA S., SHARMA G.C., 2011 - Development of new candidate gene and EST-based molecular markers for Gossypium species. - Int. J. Plant Genomics, 2011: 894598. DOI: https://doi.org/10.1155/2011/894598
- CAO Y., TANG X., GIOVANNONI J., XIAO F., LIU Y., 2012 - Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. - BMC Plant Biol., 12: 211. DOI: https://doi.org/10.1186/1471-2229-12-211
- CENCI A., GUIGNON V., ROUX N., ROUARD M., 2014 - Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. - Plant Mol. Biol., 85(1-2): 63-80. DOI: https://doi.org/10.1007/s11103-013-0169-2
- COSGROVE D.J., 2016 - Catalysts of plant cell wall loosening. - F1000Research, 5(F1000 Faculty Rev.): 119. DOI: https://doi.org/10.12688/f1000research.7180.1
- DENG G., HUANG X., XIE L., TAN S., GBOKIE T. Jr., BAO Y., XIE Z., YI K., 2019 - Identification and expression of SAUR genes in the CAM plant agave. - Genes, 10(7): 555. DOI: https://doi.org/10.3390/genes10070555
- DING A., MAROWA P., KONG Y., 2016 - Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). - Mol. Gen. Genomics, 291(5): 1891-1907. DOI: https://doi.org/10.1007/s00438-016-1226-8
- DIVAKAR S., JHA R.K., KAMAT D.N., SINGH A., 2023 - Validation of candidate gene-based EST-SSR markers for sugar yield in sugarcane. - Front. Plant Sci., 14: 1-9. DOI: https://doi.org/10.3389/fpls.2023.1273740
- DJAFARI PETROUDY S., 2017 - Physical and mechanical properties of natural fibers, pp. 59-83. - In: FAN M., and F. FU (eds.) Advanced high strength natural fibre composites in construction. Woodhead Publishing, Elsevier, Amsterdam, The Netherlands, pp. 577. DOI: https://doi.org/10.1016/B978-0-08-100411-1.00003-0
- DOYLE J.J., DOYLE J.L., 1987 - A rapid DNA isolation procedure for small quantities of fresh leaf tissue. - Phytochem. Bull., 19: 11-15.
- DUBEY L., PRASANNA B.M., RAMESH B., 2009 - Analysis of drought tolerant and susceptible maize genotypes using SSR markers tagging candidate genes and consensus QTLs for drought tolerance. - Indian J. Gen. Plant Breed., 69(4): 344-351.
- FAO, 2009 - International year of natural fibers 2009. - FAO, Food and Agriculture Organization http://www.fao.org/natural-fibres-2009/about/15-natural-fibres/en/.
- FRANCIS R.M., 2017 - Pophelper: An R package and web app to analyse and visualize population structure. - Mol. Ecol. Resour., 17: 27-32. DOI: https://doi.org/10.1111/1755-0998.12509
- GALVEZ L., KOH R., BARBOSA C., ASUNTO J., CATALLA J., ATIENZA R., COSTALES K., AQUINO V., ZHANG D., 2021 - Sequencing and de novo assembly of Abaca (Musa textilis Nee) var. Genome. - Genes, 12(8): 1202. DOI: https://doi.org/10.3390/genes12081202
- GAO C., CHENG C., ZHAO L., YU Y., TANG Q., XIN P., LIU T., YAN Z., GUO Y., ZANG G., 2018 - Genome‐wide expression profiles of hemp (Cannabis sativa L.) in response to drought stress. - Inter. J. Gen., 2018(1): 3057272. DOI: https://doi.org/10.1155/2018/3057272
- GRITSCH C., WAN Y., MITCHELL R.A., SHEWRY P.R., HANLEY S.J., KARP A., 2015 - G-fibre cell wall development in willow stems during tension wood induction. - J. Exper. Bot., 66(20): 6447-6459. DOI: https://doi.org/10.1093/jxb/erv358
- HARMER S., ORFORD S., TIMMIS J., 2002 - Characterisation of six alpha-expansin genes in Gossypium hirsutum (upland cotton). - Mol. Gen. Genomics, 268(1): 1‐9. DOI: https://doi.org/10.1007/s00438-002-0721-2
- HE Q., YU Y., QIN Z., DUAN Y., LIU H., LI W., SONG X., ZHU G., SHANG X., GUO W., 2025 - COBRA-LIKE 9 modulates cotton cell wall development via regulating cellulose deposition. - Plant Physiology, 197(1): 675. DOI: https://doi.org/10.1093/plphys/kiae675
- HUANG C., SHEN C., WEN T., GAO B., ZHU D., LI X., AHMED M.M., LI D., LIN Z., 2018 - SSR-based association mapping of fiber quality in upland cotton using an eight-way MAGIC population. - Mol. Gen. Genomics, 293: 793-805. DOI: https://doi.org/10.1007/s00438-018-1419-4
- HUANG X., BAO Y., WANG B.O., LIU L., CHEN J., DAI L., BALOCH S.U., PENG D., 2016 - Identification of small auxin-up RNA (SAUR) genes in Urticales plants: mulberry (Morus notabilis), hemp (Cannabis sativa) and ramie (Boehmeria nivea). - J. Gen., 95: 119-129. DOI: https://doi.org/10.1007/s12041-016-0622-5
- KOZIEL S.P., 2010 - Genetic analysis of lignification and secondary wall development in bast fibers of industrial hemp (Cannabis sativa).
- KULUEV B.R., KNYAZEV A.V., MIKHAYLOVA E.V., CHEMERIS A.V., 2017 - The role of expansin genes PtrEXPA3 and PnEXPA3 in the regulation of leaf growth in poplar. - Russian J. Genetics, 53(6): 651-660. DOI: https://doi.org/10.1134/S1022795417060084
- LAZAR I., LAZAR I. Jr., 2023 - GelAnalyzer 23.1.1 (available at www.gelanalyzer.com).
- LEE Y., CHOI D., KENDE H., 2001 - Expansins: ever-expanding numbers and functions. - Curr. Opin. Plant Biol., 4(6): 527-532. DOI: https://doi.org/10.1016/S1369-5266(00)00211-9
- LI F., LEE Y., KWON S., LI G., PARK Y., 2014 - Analysis of genetic diversity trait correlations among Korean landrace rice (Oryza sativa L). - Gen. Mol. Res., 13(3): 6316-6331. DOI: https://doi.org/10.4238/2014.April.14.12
- LI P., LIU Y., TAN W., CHEN J., ZHU M., LV Y., LIU Y., YU S., ZHANG W., CAI H., 2019 - Brittle Culm 1 Encodes a COBRA-Like protein involved in secondary cell wall cellulose biosynthesis in Sorghum. - Plant Cell Physiol., 60(4): 788-801. DOI: https://doi.org/10.1093/pcp/pcy246
- LIN Z., WANG Y., ZHANG X., ZHANG J., 2012 - Functional markers for cellulose synthase and their comparison to SSRS in cotton. - Plant Mol. Biol. Reporter, 30(5): 1270-1275. DOI: https://doi.org/10.1007/s11105-012-0432-8
- LIPKA A.E., TIAN F., WANG Q., PEIFFER J., LI M., BRADBURY P.J., GORE M.A., BUCKLER E.S., ZHANG Z., 2012 - GAPIT: Genome association and prediction integrated tool. - Bioinformatics, 28(18): 2397-2399. DOI: https://doi.org/10.1093/bioinformatics/bts444
- LITT M., LUTY J.A., 1989 - A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle acting gene. - Am. J. Hum. Genet., 44(3): 397-401.
- LUO Z., BROCK J., DYER J.M., KUTCHAN T., SCHACHTMAN D., AUGUSTIN M., GE Y., FAHLGREN N., ABDEL-HALEEM H., 2019 - Genetic diversity and population structure of a Camelina sativa spring panel. - Front. Plant Sci., 10: 184. DOI: https://doi.org/10.3389/fpls.2019.00184
- MALEKI S.S., MOHAMMADI K., JI K.S., 2016 - Characterization of cellulose synthesis in plant cells. - Scientific World J., 2016(1): 1-8. DOI: https://doi.org/10.1155/2016/8641373
- MATTHIES I.E., VAN HINTUM T., WEISE S., RÖDER M.S., 2012 - Population structure revealed by different marker types (SSR or DArT) has an impact on the results of genome-wide association mapping in European barley cultivars. - Mol. Breeding, 30(2): 951-966. DOI: https://doi.org/10.1007/s11032-011-9678-3
- MEHTA G., MUTHUSAMY S., SINGH G., SHARMA P., 2021 - Identification and development of novel salt-responsive candidate gene based SSRs (cg-SSRs) and MIR gene based SSRs (mir-SSRs) in bread wheat (Triticum aestivum). - Sci. Rep., 11: 2210. DOI: https://doi.org/10.1038/s41598-021-81698-3
- MENDOZA M.R.R., LAURENA A.C., DIAZ M.G.Q., OCAMPO E.T.M., LAUDE T.P., LALUSIN A.G., 2024 - Newly developed genomic SSR markers revealed the population structure and genetic characteristics of abaca (Musa textilis Nee). - BioTechnologia, 105(4): 337-353. DOI: https://doi.org/10.5114/bta.2024.145255
- MOKSHINA N., GORSHKOV O., GALINOUSKY D., GORSHKOVA T., 2020 - Genes with bast fiber-specific expression in flax plants. Molecular keys for targeted fiber crop improvement. - Ind. Crops Prod., 152: 112549. DOI: https://doi.org/10.1016/j.indcrop.2020.112549
- MORENO L., PROTACIO C., 2012 - Chemical composition and pulp properties of abaca (M. textilis Nee) cv. Inosa harvested at different stages of stalk maturity. - Visayas State University. Ann. Trop. Res., 34(2): 45-62. DOI: https://doi.org/10.32945/atr3423.2012
- NAG S., MANDAL R., MITRA J., 2020 - Exploration of genetic structure and association mapping for fibre quality traits in global flax (Linum usitatissimum L.) collections utilizing SSRs markers. - Plant Gene, 24: 100256. DOI: https://doi.org/10.1016/j.plgene.2020.100256
- NIE X., HUANG C., YOU C., LI W., ZHAO W., SHEN C., ZHANG B., WANG H., YAN Z., DAI B., WANG M., 2016 - Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. - BMC genomics, 17: 1-16. DOI: https://doi.org/10.1186/s12864-016-2662-x
- NIU E., SHANG X., CHENG C., BAO J., ZENG Y., CAI C., DU X., GUO W., 2015 - Comprehensive analysis of the COBRA-like (COBL) gene family in Gossypium identifies two COBLs potentially associated with fiber quality. - PLoS ONE, 10(12): 1-21. DOI: https://doi.org/10.1371/journal.pone.0145725
- PALUMBO F., BARCACCIA G., 2018 - Critical aspects on the use of microsatellite markers for assessing genetic identity of crop plant varieties and authenticity of their food derivatives. - In. GRILLO O. (ed.) Rediscovery of landraces as a resource for the future. IntechOpen, London, UK, pp. 204. DOI: https://doi.org/10.5772/intechopen.70756
- PRADHAN A.K., VEMIREDDY L.N.R., TANTI B., 2023 - Assessment of the genetic variability and population structure in boro rice cultivars of Assam, India using candidate gene based SSR markers. Genet. Resour Crop Evol., 70: 1747-1765. DOI: https://doi.org/10.1007/s10722-022-01533-0
- PRITCHARD J.K., STEPHENS M., DONELLY P., 2000 - Inference of population structure using multilocus genotype data. - Genetics, 155: 945-959. DOI: https://doi.org/10.1093/genetics/155.2.945
- PRITESH S.R., NAYAK N., SAMANTA S., CHHOTARAY A., MOHANTY S., DHUA S., DHUA U., PATRA B., TIWARI K.K., AMITHA S., MITHRA C., SAH R.P., BEHERA L., MOHAPATRA T., 2023 - Assessment of allelic and genetic diversity, and population structure among farmers’ rice varieties using microsatellite markers and morphological traits. - Gene Reports, 30: 10.1016. DOI: https://doi.org/10.1016/j.genrep.2022.101719
- PUTRANTO R.A., MARTIANSYAH I., SAPTARI R.T., 2017 - In silico identification and comparative analysis of Hevea brasiliensis COBRA gene family. - Proc. Int. Conf. Sci. Engin., 1: 39-47. DOI: https://doi.org/10.14421/icse.v1.267
- QIN H., CHEN M., YI X., BIE S., ZHANG C., ZHANG Y., LAN J., MENG Y., YUAN Y., JIAO C., 2015 - Identification of associated SSR markers for yield component and fiber quality traits based on frame map and upland cotton collections. - PLOSone, 10(1): e0118073. DOI: https://doi.org/10.1371/journal.pone.0118073
- RADHAKRISHNAN S., 2014 - Roadmap to sustainable textiles and clothing. Issue October 2019.
- REAMILLO M.C.S., 2018 - Genetic characterization of backcross progenies BC1, BC2 from Musa textilis Nee (Abuab) x Musa balbisiana colla (Pacol) through transcriptomic analysis. The University Library, University of the Philippine Los Baños, Laguna, Philippines.
- SAGWAL V., SIHAG P., SINGH Y., MEHLA S., KAPOOR P., BAYLAN P., KUMAR A., MIR R.R., DHANKER O.P., KUMAR U., 2022 - Development and characterization of nitrogen and phosphorus use efficiency responsive genic and miRNA derived SSR markers in wheat. - Heredity, 128: 391-401. DOI: https://doi.org/10.1038/s41437-022-00506-4
- SAH R.P., NAYAK A.K., CHANDRAPPA A., BEHERA S., AZHARUDHEEN M.TP., LAVANYA G.R., 2023 - cgSSR marker-based genome-wide association study identified genomic regions for panicle characters and yield in rice (Oryza sativa L.). - J. Sci. Food Agric., 103(2): 720-728. DOI: https://doi.org/10.1002/jsfa.12183
- SAMPEDRO J., COSGROVE D., 2005 - The expansin superfamily. - Genome Biol., 6(12): 242. DOI: https://doi.org/10.1186/gb-2005-6-12-242
- SANDOVAL C.M.C., 2011 - Molecular and biochemical studies of cassava (Manihot esculenta Crantz): seed coat phenolics, seed storage proteins and DNA fingerprinting for storage root color, hydrocyanic acid (HCN) content and bacterial blight resistance. - The University Library, University of the Philippine Los Baños, Laguna, Philippines.
- SARAGIH S., WIRJOSENTONO B., MELIANA Y., 2020 - Thermal and morphological properties of cellulose nanofiber from pseudostem fiber of abaca (Musa textilis Nee). - Macromolecular Symposia, 391(1): 2000020. DOI: https://doi.org/10.1002/masy.202000020
- SCHINDELMAN G., MORIKAMI A., JUNG J., BASKIN T.I., CARPITA N.C., DERBYSHIRE P., MCCANN M.C., BENFEY P.N., 2001 - COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. - Genes Develop., 15(9): 1115-1127. DOI: https://doi.org/10.1101/gad.879101
- SCHUMACHER C., KRANNICH C.T., MALETZKI L., KÖHL K., KOPKA J., SPRENGER H., HINCHA D.K., SEDDIG S., PETERS R., HAMERA S., 2021 - Unravelling differences in candidate genes for drought tolerance in potato (Solanum tuberosum L.) by use of new functional microsatellite markers. - Genes, 12(4): 494. DOI: https://doi.org/10.3390/genes12040494
- SENA J.S., LACHANCE D., DUVAL I., NGUYEN T.T.A., STEWART D., MACKAY J., SÉGUIN A., 2019 - Functional analysis of the PgCesA3 white spruce cellulose synthase gene promoter in secondary Xylem. - Front. Plant Sci., 10(May): 1-13. DOI: https://doi.org/10.3389/fpls.2019.00626
- SINGH A.K., CHAURASIA S., KUMAR S., SINGH R., KUMARI J., YADAV M.C., SINGH N., GABA S., JACOB S., 2018 - Identification, analysis and development of salt responsive candidate gene based SSR markers in wheat. - BMC Plant Biol., 18(1): 1-15. DOI: https://doi.org/10.1186/s12870-018-1476-1
- SOREK N., SOREK H., KIJAC A., SZEMENYEI H.J., BAUER S., HÉMATY K., WEMMER D.E., SOMERVILLE C.R., 2014 - The Arabidopsis COBRA protein facilitates cellulose crystallization at the plasma membrane. - J. Biol. Chem., 289(50): 34911-34920. DOI: https://doi.org/10.1074/jbc.M114.607192
- VICTORIA F., DA MAIA L., DE OLIVEIRA A., 2011 - In silico comparative analysis of SSR markers in plants. - BMC Plant Biol., 11: 15. DOI: https://doi.org/10.1186/1471-2229-11-15
- VINARAO G., MENDOZA M., VILLA N., DELA VIÑA C., ABUSTAN M., LALUSIN A., 2019 - Morphological Characterization and SSR-based DNA fingerprinting of cassava (Manihot esculenta Crantz) Released by the National Seed Industry Council (NSIC). - Philippine Agric. Scientist, 102(4): 350-360.
- WANG Y., BIBLE P., LOGANANTHARAJ R., UPADHYAYA H.D., 2012 - Identification of SSR markers associated with height using pool-based genome-wide association mapping in sorghum. - Mol. Breeding, 30: 281-292. DOI: https://doi.org/10.1007/s11032-011-9617-3
- XIE J., LI J., JIE Y., XIE D., YANG D., SHI H., ZHONG Y., 2020 - Comparative transcriptomics of stem bark reveals genes associated with bast fiber development in Boehmeria nivea L. gaud (ramie). - BMC Genomics, 21: 1-17. DOI: https://doi.org/10.1186/s12864-020-6457-8
- YLLANO O., DIAZ M.G.Q., LALUSIN A., LAURENA A., TECSON-MENDOZA E.M., 2020 - Genetic analyses of abaca (Musa textilis Née) germplasm from its primary center of origin, the Philippines, using simple sequence repeat (SSR) markers. - Philippines Agric. Sci., 103: 311-321. DOI: https://doi.org/10.62550/CA31029020
- ZHANG M., CAO H., XI J., ZENG J., HUANG J., LI B., SONG S., ZHAO J., PEI Y., 2020 - Auxin directly upregulates GhRAC13 expression to promote the onset of secondary cell wall deposition in cotton fibers. - Front. Plant Sci., 11: 581983. DOI: https://doi.org/10.3389/fpls.2020.581983
- ZHANG M., ZENG J.Y., LONG H., XIAO Y.H., YAN X.Y., PEI Y., 2017 - Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. - Plant Cell Physiol., 58(2): 385-397. DOI: https://doi.org/10.1093/pcp/pcw203
- ZHONG R., LEE C., ZHOU J., MCCARTHY R.L., YE Z.H., 2008 - A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. - Plant Cell, 20(10): 2763-2782. DOI: https://doi.org/10.1105/tpc.108.061325
- ZHU L., JIANG B., ZHU J., XIAO G., 2022 - Auxin promotes fiber elongation by enhancing gibberellic acid biosynthesis in cotton. - Plant Biotechnol. J., 20(3):423-425. DOI: https://doi.org/10.1111/pbi.13771





